Epi-Blood Count
The content of this page has not been updated for the last three years. It is currently under maintanance.

Epi-Blood provides a novel method to accurately determine the numbers of different leukocytes – also known as white cells – in peripheral blood. This approach is based on unique DNA methylation patterns of each subset of leukocytes.
Classical methods for differential blood counts, such as automated cell counters and flow cytometry, require viable cells and are therefore limited to fresh blood samples. If blood cannot be analyzed immediately after collection, e.g. if instrumentation is not available or long shipment is needed, conventional differential blood counts are more difficult. In contrast, the Epi-Blood method is also applicable for frozen samples. In fact, it can be even be applied to samples that have been cryopreserved for years. While our method can currently only be applied for research use only, it holds the perspective for physicians to harvest small volumes of blood for long-term storage, shipment, longitudinal, and retrospective analysis.
DNA methylation is a covalent modification of cytosine residues in the genome mostly in the context of CG dinucleotides (CpG sites). During development DNAm governs cell differentiation thereby establishing cellular identity. We have identified CpG sites where DNA methylation is characteristic for granulocytes, monocytes, and lymphocytes, as well as lymphocyte subsets CD4 T cells, CD8 T cells, B cells and NK cells. By analyzing DNA methylation at these selected CpG sites, we can determine relative (%) and absolute (cells/µl) leukocyte counts (referred to as Epi-Blood-Counts) in any given liquid or frozen blood sample. The DNA methylation levels can be measured by pyrosequencing to be entered into a non-negative least square algorithm, which returns cell proportions for each sample.
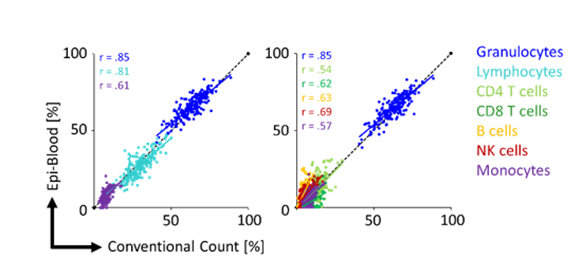
Figure 1: The relative percentage of different leukocyte subsets was compared in blood samples with an automated cell counter and the Epi-Blood method.
Absolute leukocyte quantification is possible with a reference plasmid that contains an unmethylated reference sequence and is added to each blood sample. Our work has shown good correlation (r = correlation coefficient) compared to conventional cell counts for relative Epi-Blood cell counts.
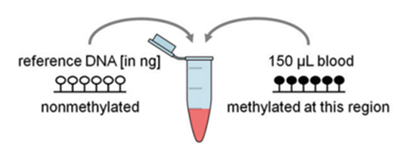
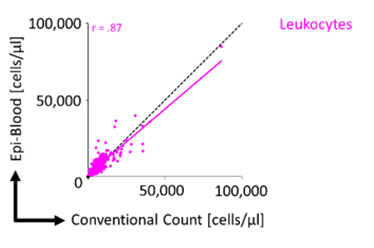
Figure 2: For absolute quantification, a non-methylated reference plasmid is added to provide relatively precise numbers of absolute cell counts.
Epi-Blood-Count is a validation project funded by the German Federal Ministry of Education and Research within the VIP+ program. We are open for further collaborations. One of the main focus of our team is the preparation of documents for acquiring the certification as in vitro diagnostic medical device (according to the new medical device regulation (EU) 2017/746) and establish a Risk and Quality Management system according to the international standards (ISO 13485 and 14971). Hereby, our method shall ultimately complement the conventional approaches for epigenetic blood counts to support better diagnostics for patients.
Supported by


